Our Research Bacterial viruses as pathogen control agents in aquaculture systems
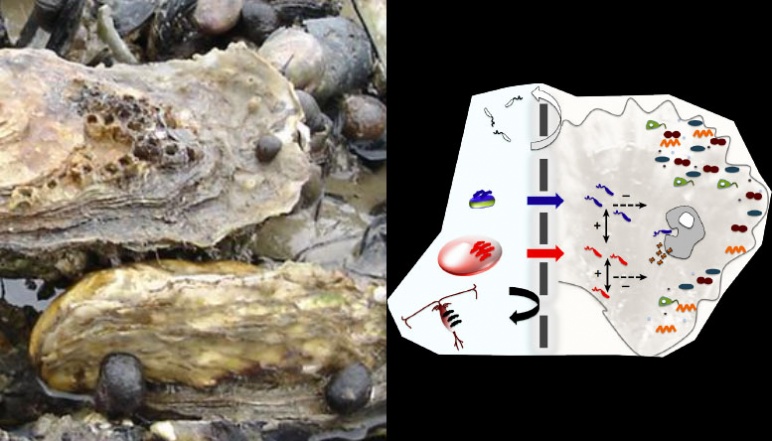
Image Credit: iStock Images, Polz research team
Principal Investigator
Martin Polz
- Former Professor
- Department of Civil & Environmental Engineering
Martin Polz is a professor with the Department of Civil & Environmental Engineering. Professor Polz’s work centers on environmental microbiology, using a combination of quantitative molecular approaches, genomics, physiology, and modeling to explore environmental and evolutionary mechanisms that trigger the emergence of pathogenic variants among microbes. He is the recipient of a 2016 J-WAFS seed grant for his project, "“Bacterial Viruses as Pathogen Control Agents in Aquaculture Systems.”
Challenge:
Can we protect cultured oysters from bacterial infection without using antibiotics?
Research Strategy
- Used bacteriophages (bacteria’s natural enemies) for pathogen control
- Used genomics to categorize viruses by host targets and assess evolution of resistance in pathogens
- Tested engineered “virus cocktails” for effectiveness in the field using naturally infected oysters
Project description
Marine aquaculture is thought to be vital for ensuring food supply for the future but like no other food production system, it is subject to catastrophic collapse due to rapidly spreading pathogens. The bacterial genus Vibrio is most commonly involved in aquaculture infections and is likely to become even more prevalent due to their positive response to warming ocean surface water. There are currently no viable treatments for vibrios in aquaculture systems since antibiotic application in aquatic systems is ineffective and causes problems due to selection for resistance. The research team in this project developed and tested bacterial viruses, bacteria’s natural enemies, as an effective and rapid treatment for pathogen infections in aquaculture systems. This approach was based on the team’s experience with vibrios and their viruses and built on an extensive collection of well-characterized isolates of both bacterial hosts and their specific viruses.
Key to finding a solution was to develop virus cocktails that target populations of genotypically diverse pathogens by combining viruses with varying host range and receptor specificity. The latter is important since it makes buildup of resistance highly improbable in the time course of treatment. Viral receptors are frequently surface proteins that play a vital role in the biology of bacteria and resistance is often associated with their mutational loss or decreased function. Hence bacteria with multiple virus resistance mutations are less competitive in the environment and should be diluted from microbial communities.
This project used the team’s existing large collection of Vibrio hosts and viruses to characterize receptors that allow different virus families to infect, verify the host range of these different virus families, and assemble and test virus cocktails for their effectiveness in killing pathogen populations in laboratory and field experiments. This proof of concept work was carried out in oysters and Vibrio pathogens that infect them through collaboration with colleagues in France where up to 90% of juvenile oysters are currently killed by V. crassostreae. The final product of this research was a tested virus cocktail that can be relatively easily be produced in large quantities and applied in the field. Because the project team worked with natural isolates, no regulatory issues of using biologically engineered organisms occurred. The protocols and logic of this approach can be broadly applied to other aquaculture and agricultural systems affected by recurrent infections of the same pathogen population.
Outcomes
- Used genome sequencing to create the largest network of bacteria-virus interactions to investigate the host range of different viruses
- Analyzed what genetic factors impact host-virus interactions
- Discovered novel mechanisms of phage resistance in bacterial hosts involving mobile genetic elements that can be integrated into host genome
- Clarified the nature of viruses to switch hosts through recombination with other viruses
Additional Details
Impact Areas
- Food
Research Themes
- Transforming Food Systems
Year Funded
- 2016
Grant Type
- Seed Grant
Status
- Completed
